Table Of Content

The program will return, if possible, only primer pairs that do not generate a valid PCR product on unintended sequences and are therefore specific to the intended template. Note that the specificity is checked not only for the forward-reverse primer pair, but also for forward-forward as well as reverse-reverse primer pairs. This is another parameter that can be used to adjust primer specificity stringecy. For examaple, if you are only interested in targets that perfectly match the primers, you can set the value to 1.
Golden Gate Cloning Method of Primer Designing
FBPP: software to design PCR primers and probes for nucleic acid base detection of foodborne pathogens Scientific ... - Nature.com
FBPP: software to design PCR primers and probes for nucleic acid base detection of foodborne pathogens Scientific ....
Posted: Fri, 12 Jan 2024 08:00:00 GMT [source]
You can also lower the E value (see advanced parameters) in such case to speed up the search as the high default E value is not necessary for detecting targets with few mismatches to primers. The nanomolar concentration of annealing oligos in the PCR. Note that this is not the concentration of oligos in the reaction mix but of those annealing to template. Primer3 uses this argument to calculate oligo melting temperatures. This parameter corresponds to 'c' in Rychlik, Spencer and Rhoads' equation (ii) (Nucleic Acids Research, vol 18, num 21) where a suitable value (for a lower initial concentration of template) is "empirically determined".
Related products and tools
Polymerization direction is into the template stand rather outward of sequence as happens without reverse complementation. Methylation Specific PCR (MSP) relies on amplification to assess the methylation status at specific CpG sites after bisulfite conversion. Success with this system depends on the differential amplification of the template using methylated (M) and non-methylated (U) primer sets. While most of the considerations for MSP primer design are identical for those for bisulfite PCR, CpG sites within the primers are treated differently. Bisulfite PCR examines the methylation status of all the CpG sites present in a specific region, while MSP assesses the methylation at a single CpG site complementary to the 3’ end of the primer sequence. Enabling this option will make it much easier to find gene-specific primers since there is no need to distinguish between splice variants.
New to the PrimerQuest Tool?
The millimolar concentration of deoxyribonucleotide triphosphate. This argument is considered only if Concentration of divalent cations is specified. The maximum number of Gs or Cs allowed in the last five 3' bases of a left or right primer.
Development of PCR primers enabling the design of flexible sticky ends for efficient concatenation of long DNA fragments - ScienceDirect.com
Development of PCR primers enabling the design of flexible sticky ends for efficient concatenation of long DNA fragments.
Posted: Wed, 03 Apr 2024 07:00:00 GMT [source]

Gibbs’s free energy plays a very pivot role in primer designing. This is because of the spontaneous reaction at constant temperature and pressure. Thereby, higher G denotes(greater than 0, or positive G) implies an enthalpy to form while secondary structures take low spontaneous reaction with lower G value. The very negative G indicates the affinity to form a structure to linear form with the release of heat in the reverse back manner thus, being more a stable secondary structure(larger negative G values) should be avoided.
NEBuilder HiFi DNA Assembly & Gibson Assembly
Illustration showing the importance of reverse complementation. 5.3 Binding of primer to the complementary strand and extension of the primer. Polymerization results in the synthesis of the upper strand (5’-3’). This action is equivalent to the action of forward primer.
Primer Designer Tool for PCR & Sanger Sequencing
The positions refer to the base numbers on the plus strand of your template (i.e., the "From" position should always be smaller than the "To" position for a given primer). Note that the position range of forward primer may not overlap with that of reverse primer. Although we have mentioned ideal conditions to make primers, it is not always possible to control the sequence selection (amplifying gene from the start codon for example).
However, the BLAST program reports a statistical significance, called “expectation value”(E – value) for each alignment which is an indicator for finding the match by chance. E – values ≤ 0.01 convey the homologous sequences (Altschul et al., 1990)( Karlin et al., 1990). E- value measures for assessing potential biological relationships (Raymaekers M et al, 2009). Despite the fact that Insilco tools provide valuable feedback, the specificity of the qPCR assay using the designed primers and probes has to be validated empirically with direct experimental evidence (Bustin et al., 2009). Bisulfite PCR is a multi-step process that enables users to determine the methylation status of CpG sites within a PCR amplicon. The first step in bisulfite PCR is bisulfite conversion of the DNA.
Also, both of the 3’ ends of the hybridized primers must point toward one another. This requires that the left or the right primers to span a junction that is just 3' of any such positions. For example, entering "50 100" would mean that the left or the right primers must span the junction between nucleotide position 50 and 51 or the junction between position 100 and 101 (counting from 5' to 3'). You can also specify in the fields below the minimal number of nucleotides that the left or the right primer must have on either side of the junctions. This option is useful if you want a primer to a span specific junction on the template. Note that this option cannot be used in association with the "Exon/intron selection" options above.
This specifies the range of total intron length on the corresponding genomic DNA that would separate the forward and revervse primers.
Cross-check if the sequence has been reverse complemented or not. Image showing the steps involved in reverse complementation in APE software. We are using APE (A Plasmid Editor) software to design the primers, which is free to use.
Choose a higher value if you need to perform more stringent search. This controls whether the primer should span an exon junction on your mRNA template. The option "Primer must span an exon-exon junction" will direct the program to return at least one primer (within a given primer pair) that spans an exon-exon junction. This is useful for limiting the amplification only to mRNA. You can also exclude such primers if you want to amplify mRNA as well as the corresponding genomic DNA.
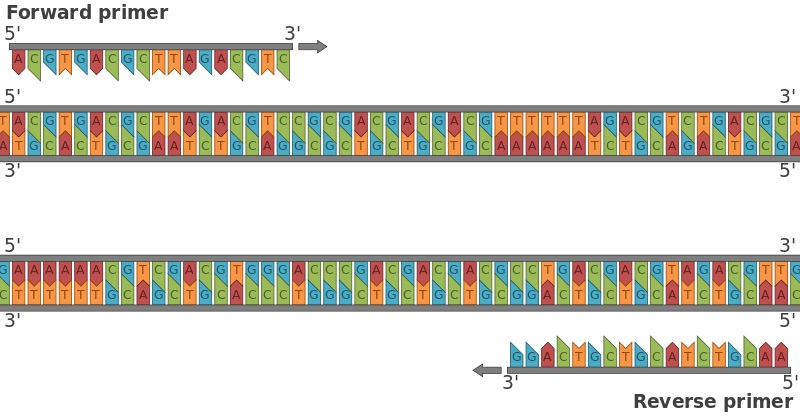
Apart from the PCR, DNA sequencing primers combine with restriction cloning, as well as other DNA new assemblies such as Gibson DNA assembly methods together with Golden Gate method. The modified step annealing can be performed using gradient PCR where temperature can be set to bind primers. Forward and Reverse primers don’t follow the complementarity rule, rather a forward primer binds to one end of one target at 5’ P while the other end of 5’ P occupies reverse primer. The forward primer runs in 3’-5’ while the reverse primer runs in 5’-3’. However process of elongation results in two new strands of ds DNA.

No comments:
Post a Comment